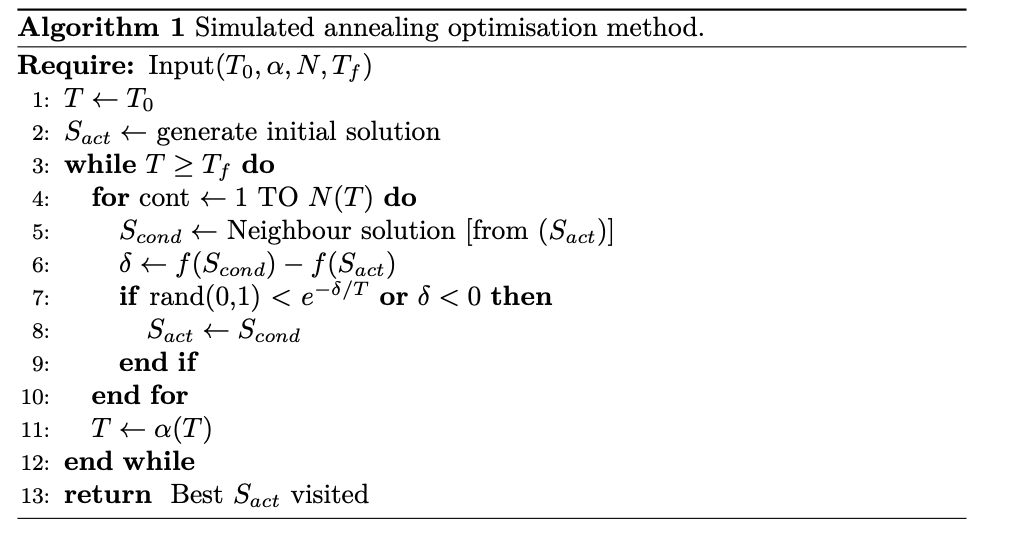
1. Simulated annealing optimisation method🔗
np.random.seed(55) # For reproducibility
# Section 3.d.1
def eggHolder(x, y):
return -(y + 47)*np.sin(np.sqrt(np.abs(x/2 + (y + 47)))) - x * np.sin(np.sqrt(np.abs(x - (y+47))))
# simulated_annealing
def simulated_annealing_with_range(initT, alpha, nIter, finalT, range_x, range_y):
temp = initT
current = (np.random.uniform(range_x[0], range_x[1]), np.random.uniform(range_y[0], range_y[1])) # Random initial solution
best = current
# Store all solutions as a list of (x,y) tuples
solutions = []
solutions.append(current)
# Store temperature at each iteration
y_values = np.array([])
y_values = np.append(y_values, eggHolder(*current))
while temp >= finalT:
for _ in range(nIter):
# 自适应步长:步长随温度缩放,大温度大步探索,小温度小步精修
sigma0 = 80.0
neighbor = (
current[0] + np.random.randn() * sigma0 * (temp / initT) ** 0.7,
current[1] + np.random.randn() * sigma0 * (temp / initT) ** 0.7
)
# 反射进边界
nx = min(max(neighbor[0], range_x[0]), range_x[1])
ny = min(max(neighbor[1], range_y[0]), range_y[1])
neighbor = (nx, ny)
deltaE = eggHolder(*neighbor) - eggHolder(*current)
metropolis = np.exp(-deltaE / temp)
if deltaE < 0 or np.random.rand() < metropolis:
current = neighbor
if eggHolder(*current) < eggHolder(*best):
best = current
temp *= alpha # Cool down
# Record current solution and value
solutions.append(current)
y_values = np.append(y_values, eggHolder(*current))
# convert to numpy array shape (n_iters, 2) before returning/plotting
return best, np.array(solutions), y_values
#
best, solutions, y_values = simulated_annealing_with_range(
1000, 0.9, 1000, 1e-3, (-512, 512), (-512, 512))
print(f"Simulated Annealing Result: x = {best}, f(x) = {eggHolder(*best):.6f}")
# plotting the solutions and temperature over iterations
plt.figure(figsize=(12, 5))
plt.ylabel('Value')
plt.plot(y_values, label='Value')
plt.xlabel('Iteration')
plt.ylabel('Solution Value')
plt.show()
# plt.plot(solutions[:, 0], solutions[:, 1])
# plt.xlabel('X')
# plt.ylabel('Y')
# plt.title('Solutions over Iterations')
# plt.grid()
# plt.show()
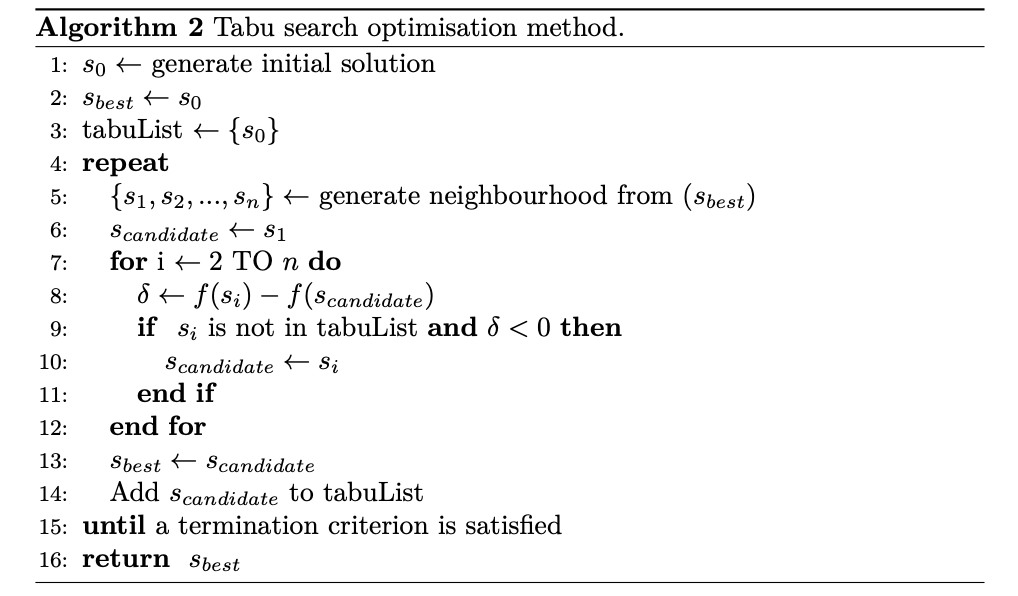
2. Tabu search optimisation method🔗
def simulated_tabu_search(neighbor_size, criteria):
init = np.random.randn() * 10 # Random initial solution
best = init
tabu_list = np.array([])
tabu_list = np.append(tabu_list, init)
while True:
neighbors = np.zeros(neighbor_size)
for j in range(neighbor_size):
neighbors[j] = best + np.random.randn() # Generate neighbors
candidate = neighbors[0]
for i in range(1, neighbor_size):
delta = f(neighbors[i]) - f(candidate)
in_tabu = any(abs(x - t) < 1e-5 for t in tabu_list)
if delta < 0 and not in_tabu:
candidate = neighbors[i]
best = candidate
tabu_list = np.append(tabu_list, best)
if f(best) < criteria:
break
return best
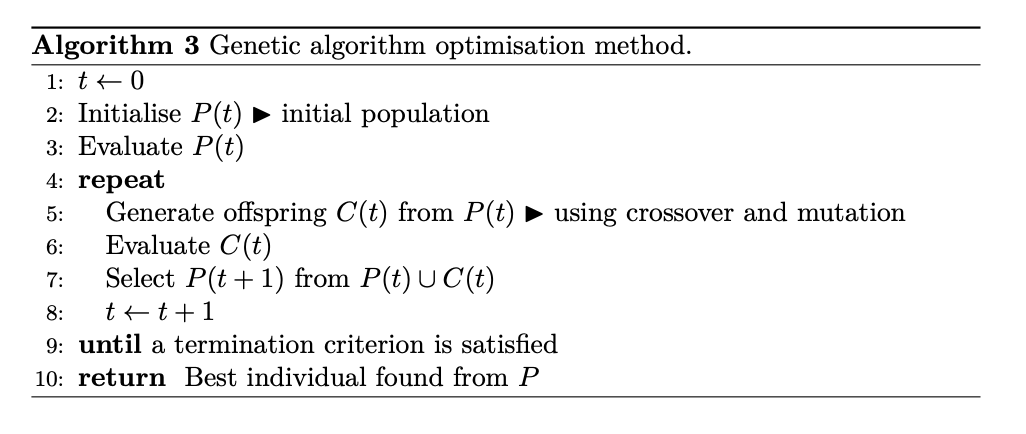
3. Genetic Algorithm🔗
"""
Created on Fri Jun 27 16:16:14 2025
@author: Francisco
"""
import random
random.seed(42) # For reproducibility
def genetic_algorithm(config):
#Parameters
TARGET = config.get("TARGET", "HELLO WORLD")
POPULATION_SIZE = config.get("POPULATION_SIZE", 100)
MUTATION_RATE = config.get("MUTATION_RATE", 0.9)
GENERATIONS = config.get("GENERATIONS", 1000)
#Generates a random string of a given length
def generate_individual(length):
return ''.join(random.choice("ABCDEFGHIJKLMNOPQRSTUVWXYZ ") for _ in range(length))
#Calculates the fitness of an individual based on how closely it matches the target
def calculate_fitness(individual):
fitness = 0
for i in range(len(TARGET)):
# if i < len(individual) and individual[i] == TARGET[i]:
if individual[i] == TARGET[i]:
fitness += 1
return fitness
#Selects two parents based on their fitness (roulette wheel selection)
def select_parents(population, fitnesses):
total_fitness = sum(fitnesses)
if total_fitness == 0: # Handle case where all fitnesses are zero
return random.sample(population, 2)
rnd1 = random.uniform(0, total_fitness)
rnd2 = random.uniform(0, total_fitness)
parent1 = None
parent2 = None
current_sum = 0
for i, individual in enumerate(population):
current_sum += fitnesses[i]
if parent1 is None and current_sum >= rnd1:
parent1 = individual
if parent2 is None and current_sum >= rnd2:
parent2 = individual
if parent1 is not None and parent2 is not None:
break
return parent1, parent2
#Performs single-point crossover between two parents
def crossover(parent1, parent2):
crossover_point = random.randint(1, len(parent1) - 1)
child1 = parent1[:crossover_point] + parent2[crossover_point:]
child2 = parent2[:crossover_point] + parent1[crossover_point:]
return child1, child2
#Mutates an individual by randomly changing characters
def mutate(individual, mutation_rate):
mutated_individual = list(individual)
for i in range(len(mutated_individual)):
if random.random() < mutation_rate:
mutated_individual[i] = random.choice("ABCDEFGHIJKLMNOPQRSTUVWXYZ ")
return "".join(mutated_individual)
def run_genetic_algorithm():
#Generate n (POPULATION_SIZE) strings of a given lenght
population = [generate_individual(len(TARGET)) for _ in range(POPULATION_SIZE)]
for generation in range(GENERATIONS):
fitnesses = [calculate_fitness(ind) for ind in population]
#Check for solution
best_fitness = max(fitnesses)
best_individual_index = fitnesses.index(best_fitness)
best_individual = population[best_individual_index]
if best_individual == TARGET:
print(f"Target found in generation {generation}: {best_individual}")
return
# print(f"Generation {generation}: Best Fitness = {best_fitness}/{len(TARGET)}, Best Individual = {best_individual}")
new_population = []
for _ in range(POPULATION_SIZE // 2): # Create pairs of children
parent1, parent2 = select_parents(population, fitnesses)
child1, child2 = crossover(parent1, parent2)
new_population.append(mutate(child1, MUTATION_RATE))
new_population.append(mutate(child2, MUTATION_RATE))
population = new_population
print("Genetic algorithm finished without finding the exact target.")
print(f"Final best individual: {best_individual}, Fitness: {best_fitness}/{len(TARGET)}")
